All ORFs marked as “cloned” by the ORFeome project can be obtained through the Centre International de Ressources Microbiennes (CIRM) .
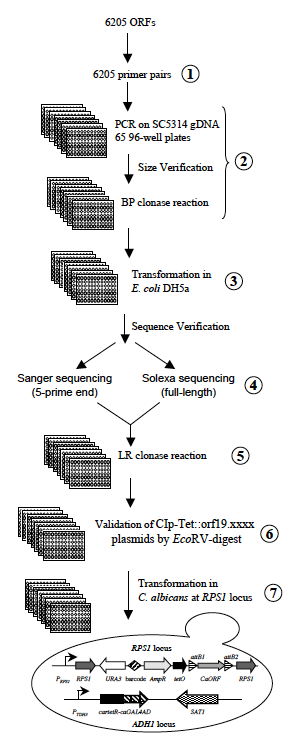
Step 2: The ORF are amplified from genomic DNA of C. albicans strain SC5314 using high fidelity polymerase. PCR products are gel verified for size and quantity and subsequently transferred into the pDONR207 donor vector using BP clonase.
Step 3: BP clonase reactions are transformed into E. coli and one transformant per ORF is stored as well as the remainder of the transformation mixture. This will allow the future identification of additional pDONR plasmids for each ORF corresponding to the second allele of the ORF.
Step 4: pDONR plasmids are individually prepared and subjected to Sanger sequencing from the 5-prime end of the ORF in order to ensure proper clone allocation in 96-well plates. Furthermore, the pDONR plasmids are subjected to Solexa sequencing and the resulting sequencing data are compared to ORF sequences obtained from CGD and SNP data generated by Solexa sequencing of C. albicans strain SC5314. These data are used to decide whether to proceed to Step 5 or reiterate the procedure, eg if mutations in the cloned ORF are not validated from Solexa sequencing data from C. albicans strain SC5314.
Step 5: ORFs in pDONR plasmids are individually transferred into a barcoded destination vector CIp10-TETp-GTW, using LR clonase, resulting in the CIp-Tet::orf19.xxxx plasmid.
Step 6: CIp-Tet::orf19.xxxx plasmids are individually prepared and gel verified following a EcoRV-digest.
Step 7: StuI or I-SceI-digested CIp-Tet::orf19.xxxx plasmids are transformed in a C. albicans strain constitutively expressing a C. albicans-adapted reverse Tet-dependent transactivator, allowing doxycycline-inducible expression of the ORF in the CIp-Tet::orf19.xxxx plasmid. Integration of the CIp-Tet::orf19.xxxx plasmid at the RPS1 locus is verified by colony PCR.
The Candida albicans ORFeome collection is made possible through the collaboration between the d’Enfert lab (Fungal Biology and Pathogenicity Unit, Institut Pasteur, Paris, France) and the Munro lab (Aberdeen Fungal Group, University of Aberdeen, Scotland).
